GROMACS version: 2021.5
GROMACS modification: No
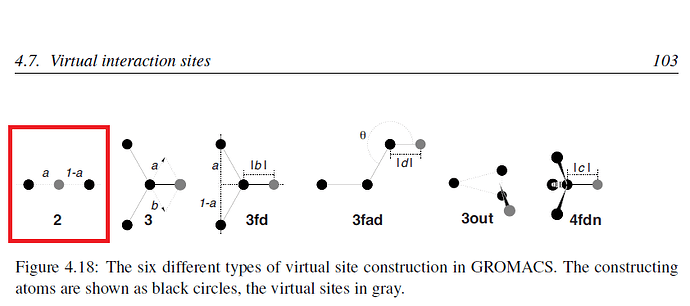
I am using a model for O2 that has one virtual site placed at d/2, where d = distance between oxygen atoms in a O2 molecule (Modelling Simul. Mater. Sci. Eng. 24 (2016) 045002). I want to be sure to correctly apply this model. This is the itp file:
[ moleculetype ]
; molname nrexcl
O2 3
[ atoms ]
; id at type res nr residu name at name cg nr charge
1 O 1 O2 O 1 -0.112
2 O 1 O2 O 1 -0.112
3 WO 1 O2 WO 1 0.224
[ bonds ]
1 2 1 0.1017 500000
[ virtual_sites2 ]
; Vsite from funct a (fraction distance)
3 1 2 1 0.5
The following is the non-bonded interactions:
[ defaults ]
; nbfunc comb-rule gen-pairs fudgeLJ fudgeQQ
1 3 yes 0.5 0.5
[ atomtypes ]
;name at.num mass charge ptype sigma epsilon
O 8 15.9990 0 A -0.112 0.42180
WO 0 0 0 D 0.224 0
These are the sections of manual I am following:
Javier Luque Di Salvo