GROMACS version: 2023
GROMACS modification: No
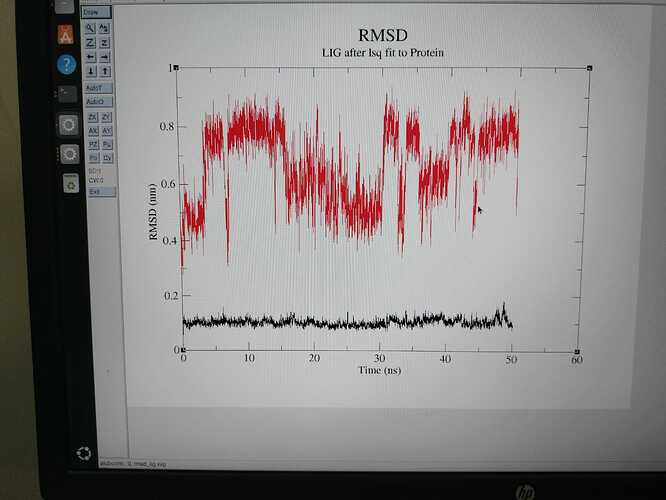
i am new to md simulation, i have done several md simulation for various protein and ligands. for all the time i am higher deviation for ligand when comparing to the protein. i don’t know whether it is right or wrong.
could anyone able to say the mistake i am doing.
thank you advance.
Hi,
You are fitting to the protein and so this is showing the RMSD of the ligand with respect to the protein and not in respect to itself. In other words, you are seeing a measure of how much the ligand is deviating from its starting position in the protein. If you want to see how much the ligand structure is deviating with regard to its own structure then you need to fit to the ligand and not the protein.
Cheers
Tom