Dear all
I had submitted an all atom md simulation for two docked(using HDOCK Server) protein stretches containing 103 and 9 residues.As per my knowledge during mindist calculation it automatically consider PBC but when I am calculating mindist between two protein chains I am getting a large jump in distance even when I am using PBC yes.Can any one suggest a solution for this ?
That’s unusual if the trajectory is normal. You might need to observe the trajectory to determine what happened at the jump point, and then try the solution based on this.
Hi Seyilaxa
As per my observation trajectory is looking fine.I have calculated mindist using the command
gmx_mpi mindist -f complex_500dt.xtc -s em.tpr -n index.ndx -od mindist_chA_chB
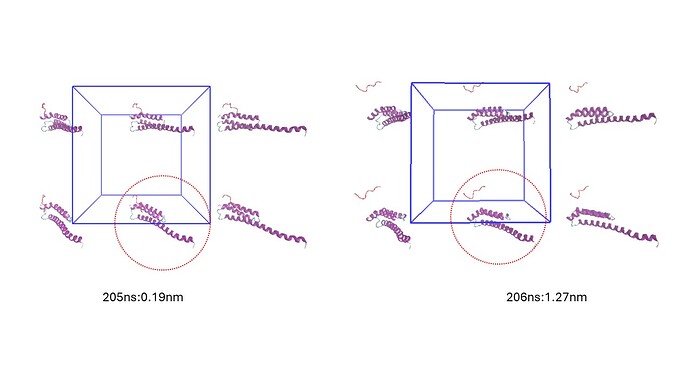
I found the first during time scale from 205ns to 206ns i.e 0.19nm to 1.27nm(snapshot attached)
I have tried following steps for pbc correction as per suggestions given by different peoples
First I tried to centerd and then fitted by these commands
gmx_mpi trjconv -s em.tpr -f complex_dt500.xtc -o centered.xtc -center -pbc mol -ur compact(selecting 1 - Protein and 0 - system for output
gmx_mpi trjconv -s em.tpr -f centered.xtc -o fitted.xtc -fit rot+trans(selecting 4 - Backbone)
By using these out I have calculated mindist between these protein chains but I found these jump again.
Then I have tried these commands
gmx_mpi trjconv -s em.tpr -f complex_dt500.xtc -o whole.xtc -pbc whole
gmx_mpi trjconv -s em.tpr -f whole.xtc -o whole_nojump -pbc nojump
gmx_mpi trjconv -s em.tpr -f whole_nojump.xtc -o whole_nojump_mol.xtc -pbc mol
Then also I didn’t get any fruitful results.
After that I have gone through this link GitHub - BartBruininks/mdvwhole: Density based object completion over PBC. and follwed the steps then also didn’t found the pbc correction and still getting such jump during mindist calculation.
I think you should show the box and determine whether the separation caused by PBC or the protein and peptide are exactly separated.
I am getting a jump of 1.08nm for a 1ns i.e from 205ns to 206ns.Is such separation for a single ns acceptable ?
It depends on your research objective. If you want to confirm the complex’s stability by MD simulation, you should at least provide a reasonable viewpoint to explain the separation like these.
I was thinking that the jump is due to PBC but it’s not because of PBC.It’s becomes closer after certain times.In literature it is found that it bounds but during MD simulation I am getting such separation.I want to check the stability of the binding of the complex.Is there any other possible reasons for this separation.
Perhaps it’s just an ordinary unbinding event. I guess the peptide is moving freely in the solution after separating from the protein. If you simulate long enough (could be very long) it may bind again.
There might be many reasons for this.
First, the simulation result largely depends on whether your initial complex structure is stable or not. Mostly, the crystall structure obtained through experment will be more credible than the docking result. So if you use the molecules docking to get the initial complex, you might need to try more docking result.
Additionally, sometimes the randomly generated speed can lead to an unstable initial strucure, so changing the seed and repeating the simulation from different initial structure may change the simulation result. Before that, make sure your forcefied is chosen reasonably to exclude the reason caused by forcefield parameters.
Finally, ligands and protein also do not always be bound in reality. Perhaps you just get the separation process. Extending the simulation time may demonstrate the stability and trend of binding from the time of binding and separation, but the time needs to be extended is unknown (maybe far beyond the time you can accept ).