GROMACS version:2021
GROMACS modification: No
Hi all,
I am simulating a system of lipids and peptides to study peptide penetration into lipids using CG-MARTINI. The input files were generated using CHARMM-GUI.
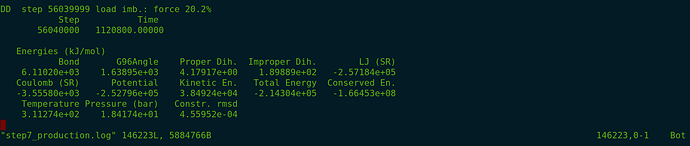
I simulate the system, and the simulation runs till a point and prints output xtc and log. It suddenly stops printing any output while the simulation is still running in the background. In the log file, there are no errors or big leaps in the energy and temperature of the system.
Please let me know if someone has faced this kind of problem. If so, what to be done?
##md.mdp file for reference
integrator = md
tinit = 0.0
dt = 0.020
nsteps = 1000000000
nstxout = 5000
nstvout = 5000
nstfout = 5000
nstlog = 5000
nstenergy = 5000
nstxout-compressed = 5000
compressed-x-precision = 1000
cutoff-scheme = Verlet
nstlist = 20
ns_type = grid
pbc = xyz
verlet-buffer-tolerance = 0.005
epsilon_r = 15
coulombtype = reaction-field
rcoulomb = 1.1
vdw_type = cutoff
vdw-modifier = Potential-shift-verlet
rvdw = 1.1
tcoupl = v-rescale
tc-grps = protein membrane solute
tau_t = 1.0 1.0 1.0
ref_t = 310 310 310
; Pressure coupling:
Pcoupl = Parrinello-rahman
Pcoupltype = semiisotropic
tau_p = 12.0
compressibility = 3e-4 3e-4
ref_p = 1.0 1.0
; GENERATE VELOCITIES FOR STARTUP RUN:
gen_vel = no
refcoord_scaling = all
#######################
Screenshot: