GROMACS version: 2018.6
GROMACS modification: No
Dear all,
I am trying to simulate a polysaccharide. I have checked that its monomer topology is present in the .rtp file. How to build its topology? Furthermore when I tried pdb2gmx it showed that some atom is missing from pdb file.[ As far as I understand this error is perhaps regarding the glycosidic bonds which is not recognized by pdb2gmx]. Thanks in advance.
Anirban
Please provide the actual error message so we can better diagnose and provide advice.
You will need to create terminal residue entries in the .rtp file for the reducing and non-reducing ends of the polysaccharide, and you should not apply terminal patching via .tdb entries, because those won’t be recognized for non-protein/non-nucleic acid species. See http://www.gromacs.org/Documentation_of_outdated_version/How-tos/Polymers for general concepts of what to do.
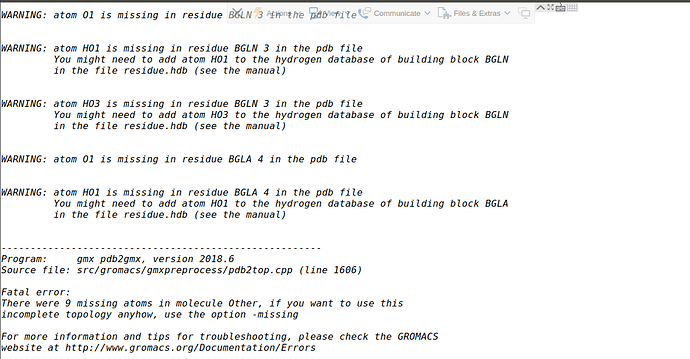
Thanks Justin @jalemkul . Here is the actual error message. BGLA and BGLN are two residues (from which I want to make my polymer) that are already present in the .rtp file.
Missing hydrogens can be rebuilt from an entry in the force field .hdb file. Other missing atoms (like O1) need to have their names corrected in your input coordinate file. All input atom names must match the force field’s expectations.
Thank you so much.I shall look into these.
Regards.
-Anirban