GROMACS version: 2025.2
GROMACS modification: No
I recently ran a 100 ns simulation on a fusion protein with flexible linkers attaching it (Glycine-Serine Linkers). The RMSD values are fluctuating in the range of 1.5 - 2.25 nm, and that’s very high. However, I’ve read that large proteins, especially flexible ones, have larger RMSD fluctuations. I visually checked how the protein conformation changes through the trajectory file input into VMD, and there was no unfolding of the structure observable, with the exception of one of the terminus loosening slightly, but otherwise the structure just moved about the linker.
Given this context, is the large RMSD values acceptable? The radius of gyration was also similar, stabilizing around 3.4 nm, but the RMSF of the linkers were approx. 1.5 angstroms, which is normal. Input would be appreciated.
Thanks.
This will depend very much on the specific case. For example, even if the biomolecule is not unfolding, if there are large fluctuations in tails or loops, you may observe large values of RMSD. Although what you report would be unusually high. Another possibility is that what you are seeing is a consequence of PBC artefacts, which sometimes become a problem for multi-chain systems. Are there discrete jumps in the RMSD?
I would suggest a careful postprocessing of the trajectory. Also, you can falsify what you see by performing the RMSD calculation of different fragments. For example: select one domain only, estimate the RMSD only for that fragment. Are the trends sensible? Also, check with alternative tools, like VMD, and see whether you get consistent results.
I hope this helps,
David
Hello,
thank you for your response! I did model the RMSD of each structure individually, and those were within much more normal ranges of 2-5 angstroms. I also viewed the trajectory through VMD, and there wasn’t any major misfolding of the structure, with the exception of one of the tails unraveling slightly.
Thank you.
Great, all of this points to problems in the RMSD calculation.
I have also experienced these things in the past. Multiple steps of postprocessing usually help solve or at least alleaviate these issues. The following series of steps has worked for me:
1. gmx trjconv -pbc cluster
2. gmx trjconv -center
3. gmx trjconv -fit rot+trans
You may have to tinker a bit with indexes, group selections and the like so that the result recapitulates what is actually happening in your trajectory.
Best,
David
Hello,
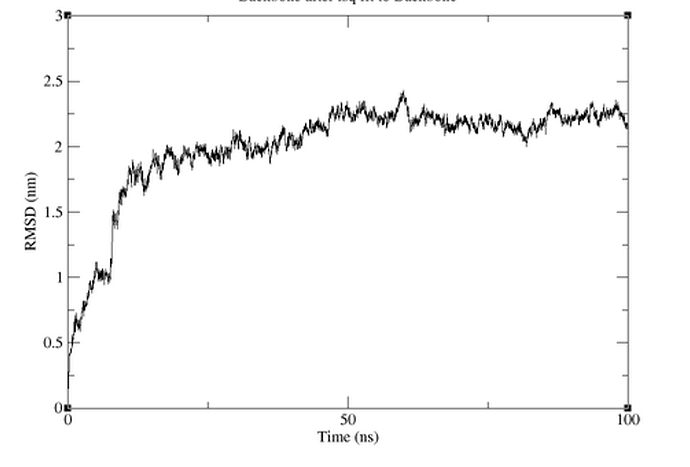
I tried multiple different combinations of indices for the commands you suggested, using protein for the fit and system for output, or protein for both, and calculating the RMSD for either the protein or the backbone, but I was getting the same graph, attached. Please let me know what you think.
Thank you.
Hmmmm, there is nothing too obvious that I can spot, sorry.
Have you tried excluding the few residues from the tail from the RMSD calculation? Does the unravelling of the tail coincide with the major jumps in RMSD (at around ~10 ns)?