GROMACS version: 2024.4
GROMACS modification: No
Hi,
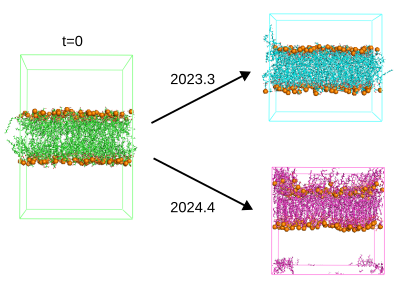
When using the deform option, in addition to weak position and dihedral restraints, to equilibrate the solvent around a lipid bilayer, I get very different outcomes, depending on whether I use gromacs 2023.3 or 2024.4. With 2023.3, the deformation proceeds well. However, the membrane gets heavily distorted when using 2024.4 (see snapshots).
I am using the same tpr in both cases, with pretty standard options:
- deform = 0 0 -0.16 0 0 0
- refcoord-scaling = COM
- position restraint along the z-axis for one head-group atom and two dihedral restraints to keep the angle between tails (as recommended by charmm-gui).
The inconsistency between versions persists if I change to a slightly different bilayer system.
Any ideas what the problem could be?
Thanks in advance,
Camilo