GROMACS version: 2020.5
GROMACS modification: Yes/No
Hello,
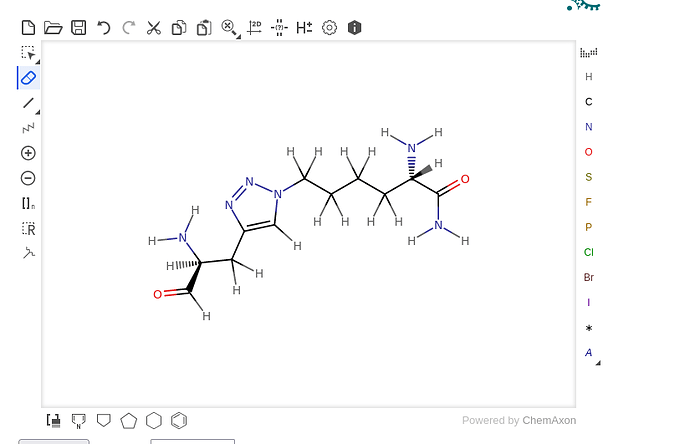
I want to perform an MD simulation using CHARMM36 ff of a stapled peptide, and it is not clear to me which procedure I should follow. I have generated parameters files of the staple using “Ligand Reader and Modeler” inside CHARMM-GUI, and I have included Calpha of the two linked residues and part of the backbones (see screenshot).
From CHARMM-GUI server I obtained .itp and .top files. How should I incorporate this information into gromacs?
The output of CHARMM-GUI is like a unique molecule, should be treated this way, or as two separate residues?