GROMACS version: 2022
GROMACS modification: No
Dear All,
I am doing MD simulation of peptides and a ligand. I used ligand reader and modeler in Charmm GUI
to generate topology files for the new ligand. When I add it with other peptides and try to run grompp (Gromacs command) the following error was generated. Can anyone suggest to me solve it ?
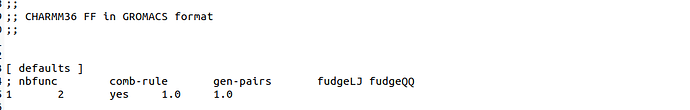
The charmm36.itp file looks like this.
Thank you !