GROMACS version: 2019. 4 and 2020.3
Hello, I use gromacs version 2019.4, but in minimization step I’ve been receiving “segmentation fault core dumped”. I’ve seen in other topics here that It would be probably a version bug that can be solved using another version, but, I’ve done my gromacs atualization and this problem is still happening. Does anyone has another answer about it?
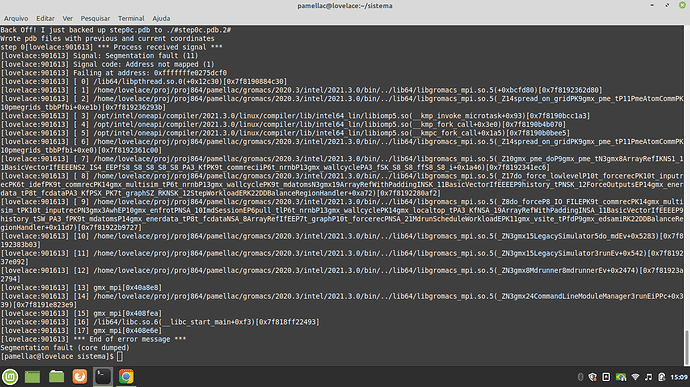
Here’s a print about what is happening in my system:
An immediate seg fault is usually a sign of infinite initial forces due to unphysical coordinates or a defective topology for some nonstandard species. You will need to provide us much more detail about what it is you’re doing to help diagnose. See Terminology — GROMACS 2021.5 documentation for general troubleshooting advice.
Thank you very much for answering me!
Well, I’m working with antibody-antigen interactions. Now I’m simulating a small epitope of the protein AMA1 from Plamodium falciparum (malaria) in water, and after building a system to simulate the complex AMA1-antibody. I’ve used ff03ws forcefield.
Simulating two proteins together shouldn’t be an issue as long as the starting coordinates are reasonable. Did grompp throw any warnings or notes? Do you have the same problem with a different force field? ff03ws is not officially supported by GROMACS, so where did you obtain the force field, and was it appropriately validated?