GROMACS version: 2022
GROMACS modification: No
Hello GROMACS community,
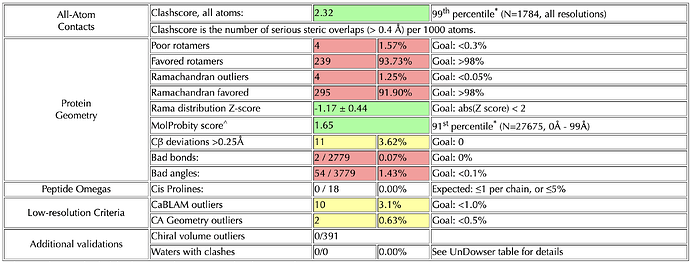
I have recently completed a simulation run, and upon inspecting my system, everything appears to be in order. However, when I analyze the protein geometry of the generated conformers using MolProbity, I encounter several severe issues suggesting improper angles, bonds, and other irregularities within my protein structure.
This discrepancy is causing me some confusion, and I would appreciate it if anyone could provide some insight into why this might be happening. I have attached the relevant outputs for reference.
Thank you for your assistance in advance.